Waveforms
Jane's second pillar besides its plugin-based document database is the
ability to deal with seismic waveforms. There is no reason to store that
information in a document database as it is very well structured; something a
relational database is made for. Thus waveform data is stored in a couple of
tables in the underlying PostgreSQL database.
Indexing Waveforms
Jane does not store the waveform in the database but only indexes some
information about it - thus you must not move data after it has been
indexed, otherwise Jane will no longer be able to find it.
To index waveforms, use the index_waveform manage.py command. As all
manage.py commands, this has to be run from the jane/src directory. It
is quite a powerful command and can recursively index a whole directory in a
single shot as well as continuously monitor a directory and automatically
index all changed files.
Usage Examples
(1.) Run only once and remove duplicates::
DATA=/path/to/archive/2015,/path/to/archive/2016
LOG=/path/to/indexer.log
python manage.py index_waveforms --verbose -i0.0 --run-once \
--check-duplicates -n1 -d$DATA
(2.) Run indexer as a daemon continuously crawling the given paths but index only the last 24 hours (-r24) of a waveform archive::
DATA=/path/to/archive/2015,/path/to/archive/2016
LOG=/path/to/indexer.log
python manage.py index_waveforms --verbose -i0.0 -n1 -d$DATA -r24 -l$LOG &
There are a lot more options, please refer to the --help output for the
most up-to-date information.
$ python manage.py index_waveforms --help
usage: manage.py index_waveforms [-h] [--version] [-v {0,1,2,3}]
[--settings SETTINGS]
[--pythonpath PYTHONPATH] [--traceback]
[--no-color] [-d DATA] [-n NUMBER_OF_CPUS]
[-i POLL_INTERVAL] [-r RECENT] [-l LOG] [-a]
[-1] [--check-duplicates] [--cleanup] [-f]
[-H HOST] [-p PORT]
Crawl directories and index waveforms to Jane.
optional arguments:
-h, --help show this help message and exit
--version show program's version number and exit
-v {0,1,2,3}, --verbosity {0,1,2,3}
Verbosity level; 0=minimal output, 1=normal output,
2=verbose output, 3=very verbose output
--settings SETTINGS The Python path to a settings module, e.g.
"myproject.settings.main". If this isn't provided, the
DJANGO_SETTINGS_MODULE environment variable will be
used.
--pythonpath PYTHONPATH
A directory to add to the Python path, e.g.
"/home/djangoprojects/myproject".
--traceback Raise on CommandError exceptions
--no-color Don't colorize the command output.
-d DATA, --data DATA Path, search patterns and feature plug-ins of waveform
files. The indexer will crawl recursively through all
sub-directories within each given path. Multiple paths
have to be separated with a comma, e.g.
'/first/path=*.*,/second/path,/third/path=*.gse'. File
patterns are given as space-separated list of
wildcards after a equal sign, e.g. '/path=*.gse2
*.mseed,/second/path=*.*'. Default path option is
'data=*.*'.
-n NUMBER_OF_CPUS Number of CPUs used for the indexer.
-i POLL_INTERVAL, --poll-interval POLL_INTERVAL
Poll interval for file crawler in seconds (default is
0).
-r RECENT, --recent RECENT
Index only recent files modified within the given
number of hours. This option is deactivated by
default.
-l LOG, --log LOG Log file name. If no log file is given, stdout will be
used.
-a, --all-files The indexer will automatically skip paths or files
starting with a dot. This option forces parsing of all
paths and files.
-1, --run-once The indexer will parse through all given directories
only once and quit afterwards.
--check-duplicates Checks for duplicate entries within database. This
feature will slow down the indexer progress.
--cleanup Clean database from non-existing files or paths if
activated, but will skip all paths marked as archived
in the database.
-f, --force-reindex Reindex existing index entry for every crawled file.
-H HOST, --host HOST Server host name. Default is 'localhost'.
-p PORT, --port PORT Port number. If not given a free port will be picked.
FDSN dataselect service
The most common way to retrieve waveforms from Jane will be via its fdsnws
dataselect service implementation. All indexed waveforms can be queried
with it. It can be found at JANE_ROOT/fdsnws/dataselect/1/ and used with
any of the common tools.
A very convenient tool it the FDSN web service client of the
ObsPy library
(documentation),
usage example with Jane:
>>> import obspy
>>> from obspy.clients.fdsn import Client
>>> c = Client("http://JANE_ROOT")
>>> st = c.get.get_waveforms(
... network="BW", station="RJOB", location="", channel="BHZ",
... starttime=obspy.UTCDateTime(2016, 1, 1, 3, 0, 5),
... endtime=obspy.UTCDateTime(2016, 1, 1, 5, 0, 5))
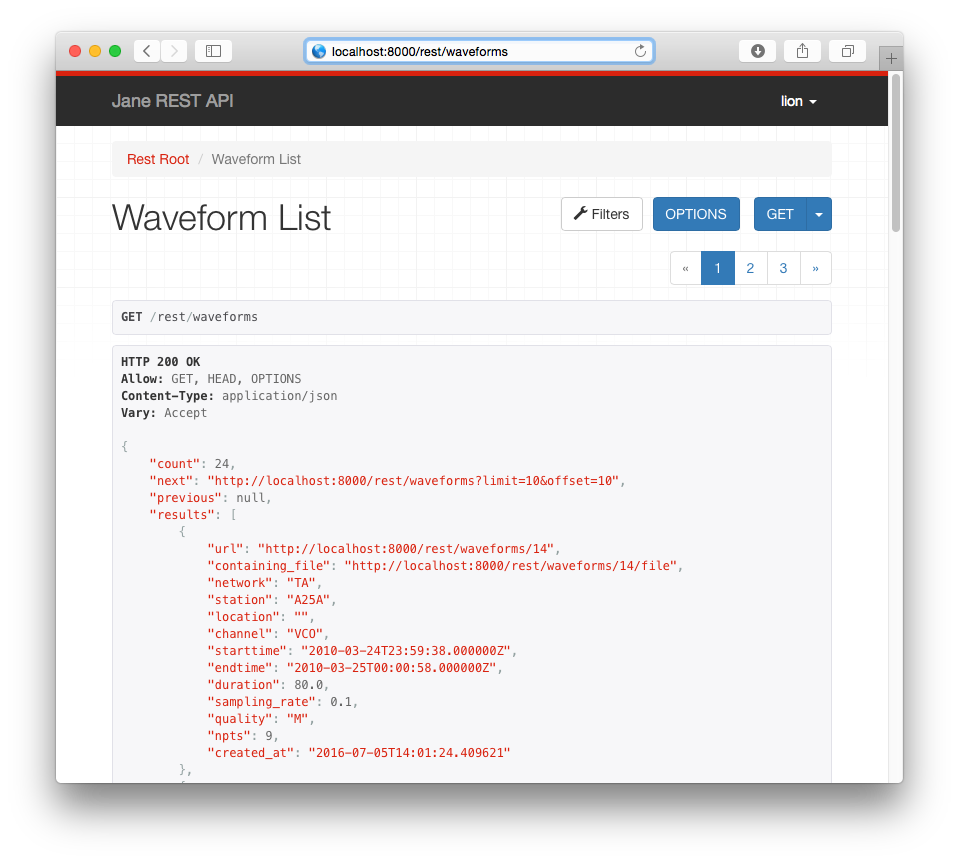
Waveform REST Interface
Waveforms can also be retrieved via the REST interface, found at
JANE_ROOT/rest/waveforms. In most cases the fdsnws service will be more
convenient but the REST interface exists if somebody needs it. It should be
fairly self-explanatory.
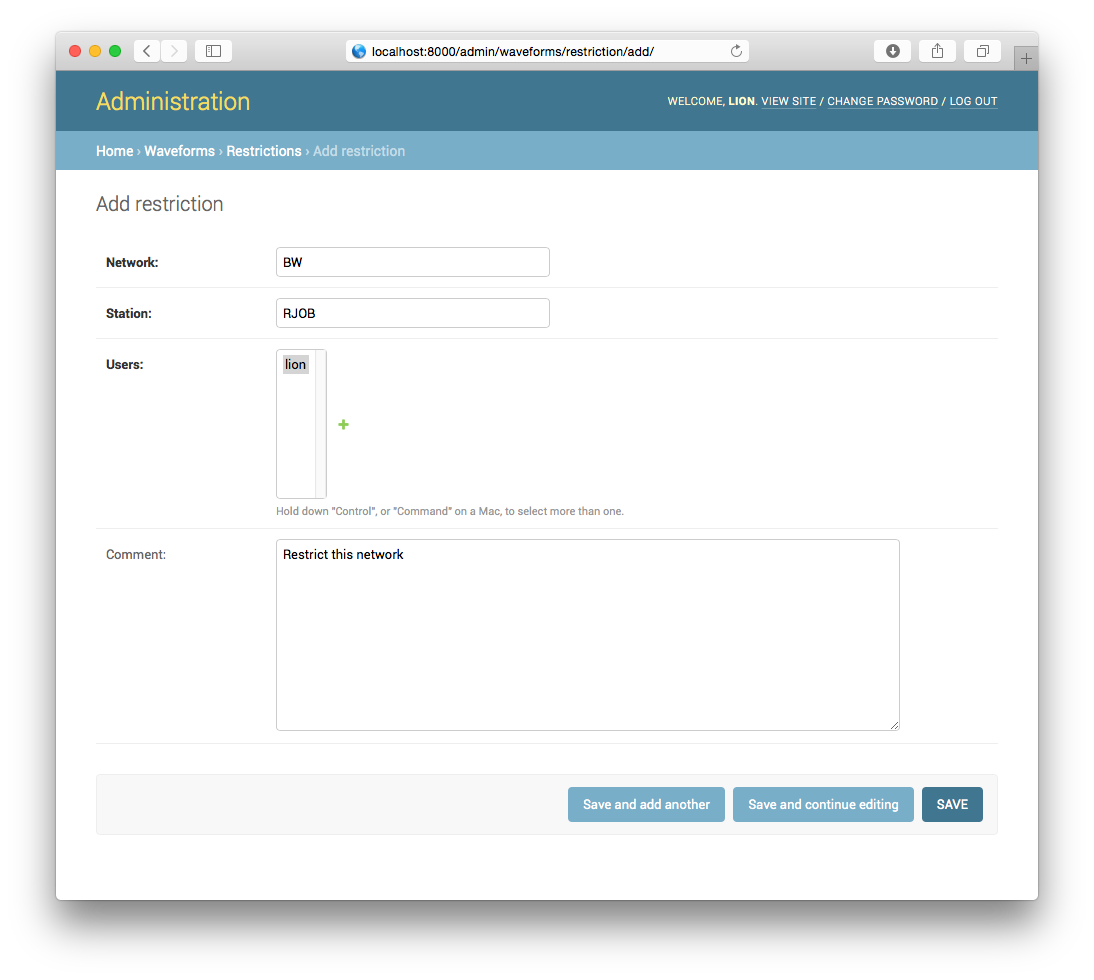
Restrictions/Protected Stations
By default all waveform data is public, i.e. anybody with access to the Jane
HTTP server can query all data. It is possible to limit access at a
per-station granularity. To do that, add a new restriction in the admin
interface. As soon as a restriction has been added it will be considered
protected and only users that are part of the restriction will still be able
to access them.
To access the data, users will have to use the queryauth route of the
fdsnws dataselect service. Usage example with ObsPy:
>>> import obspy
>>> from obspy.clients.fdsn import Client
>>> c = Client("http://JANE_ROOT", user="lion", password="myfavoritepw")
>>> st = c.get.get_waveforms(
... network="BW", station="RJOB", location="", channel="BHZ",
... starttime=obspy.UTCDateTime(2016, 1, 1, 3, 0, 5),
... endtime=obspy.UTCDateTime(2016, 1, 1, 5, 0, 5))
Waveform Mappings
Some data, especially temporary deployments, might have different network, station, location, and/or channel codes than you would like them to have. Waveform mappings to the rescue! Please note that the mappings only apply to waveforms and not the StationXML data! Add a mapping by using the admin interface:
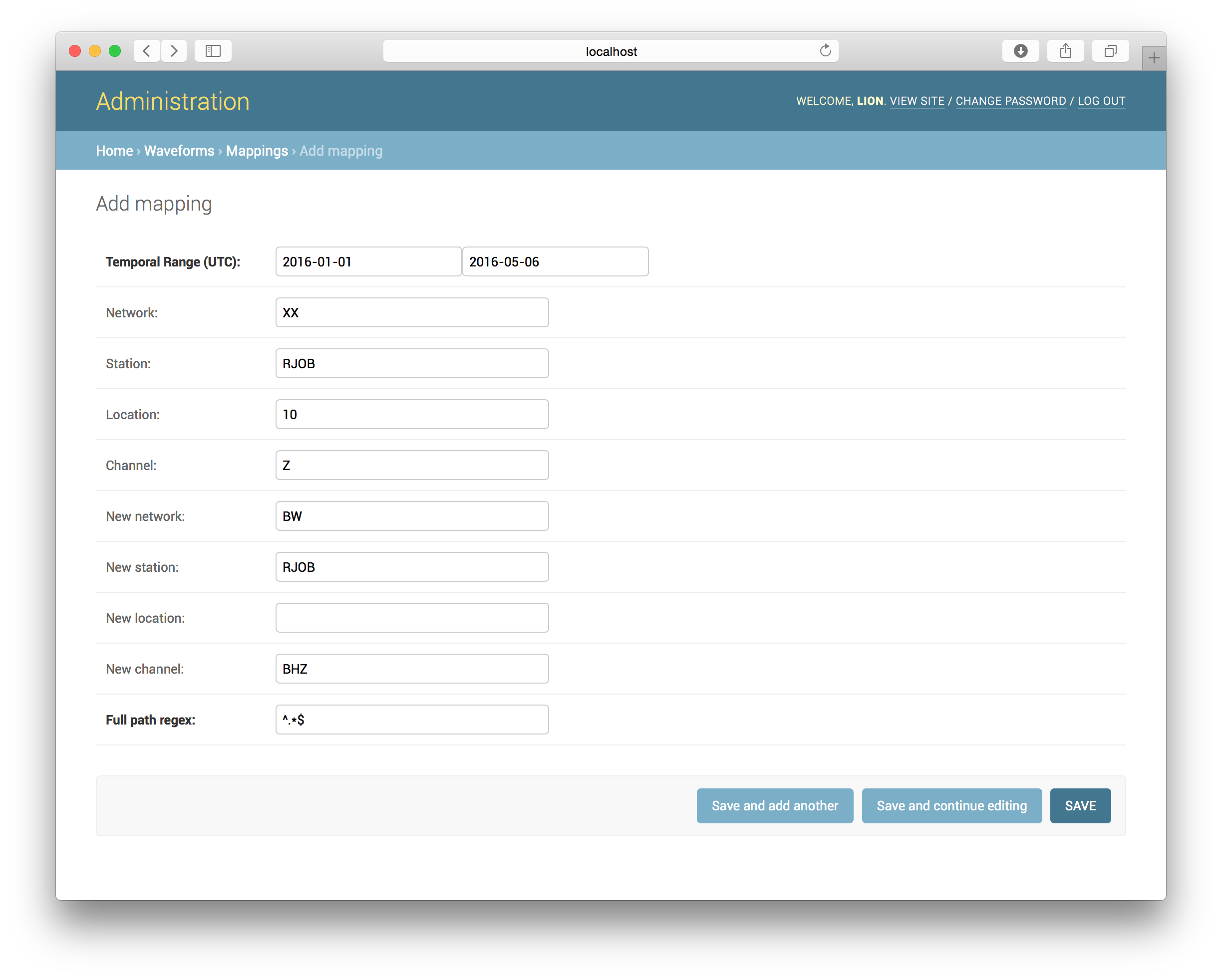
As can be seen it maps a network, station, location, channel tuple to a new
one, valid across a certain time range. Additionally, to solve the tricky
cases, each mapping takes a regular expression for its path on the filesystem.
The default value simply matches all paths. Be careful to not define
overlapping mappings - it will cause errors during the waveform indexing
process. Jane has some checks in place to prevent that but in cases
involving different regular expressions for the paths it cannot tell.
If you have a large number of mappings to apply, consider using the
add_documents management command as documented
here.
Any freshly added mapping will be automatically applied to newly indexed
data. To also apply it to existing data, press the UPDATE WAVEFORM INDICES
WITH MAPPINGS (SLOW!) button in the mappings panel in the admin interface:

This might take a while for big databases.